Overview
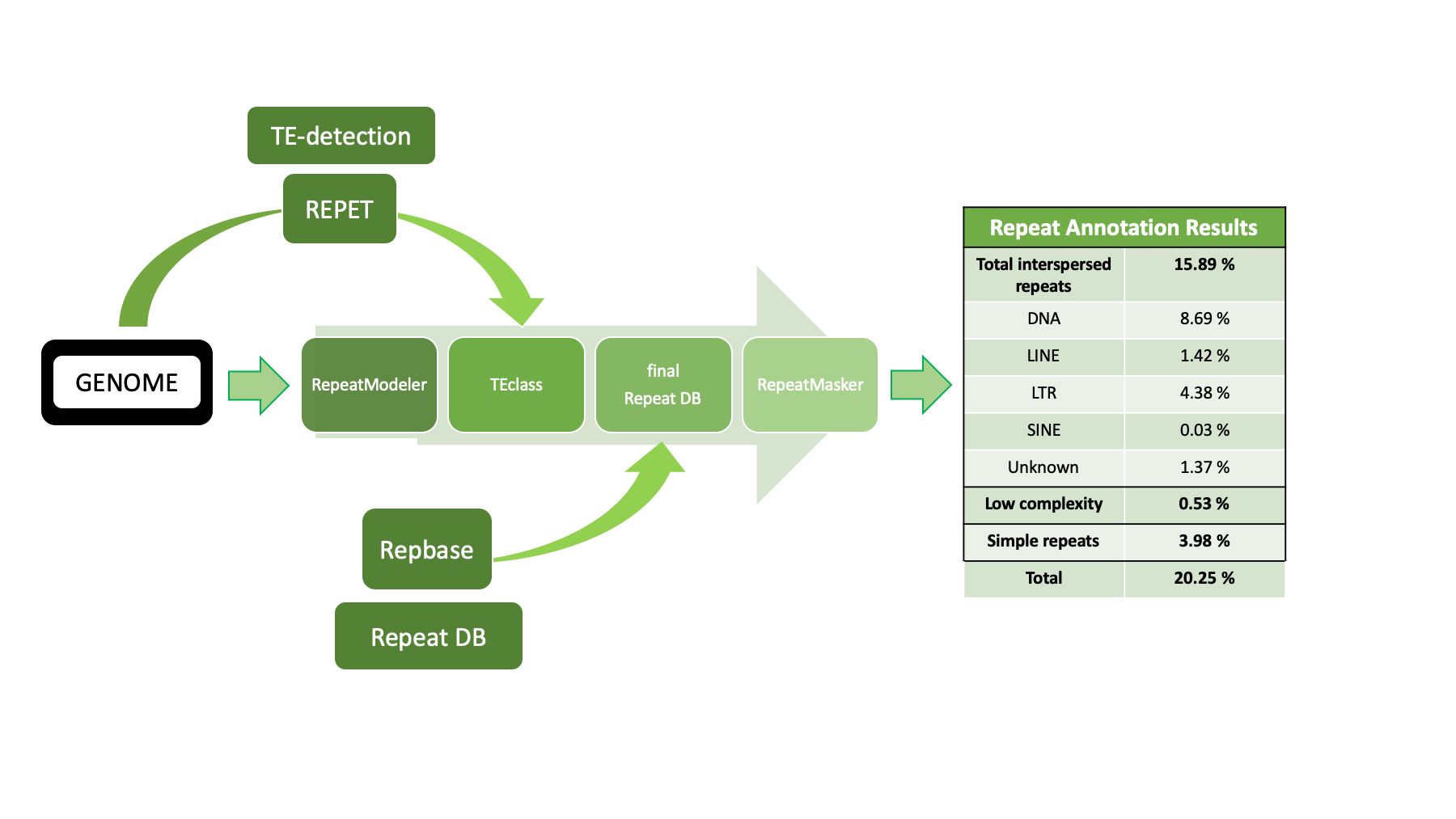
- Figure 1: Overview of the Repeat annotation pipeline with programms for identification of TE-elements (REPET) and classification of unknown repeats (TE-class).
De-novo repeat family identification with RepeatModeler
RepeatModeler builds a database for the programs out of the Genome at the first step
#Build database for RepeatModeler perl BuildDatabase -name Pcal_DB -engine ncbi P.cal_polished_new_assembly.fasta >& buildDatabase_Pcal_assembly.log
This Database is then used to build, refine, and classify consensus models of putative interspersed repeats with RECON and RepeatScoud.
#run RepeatModeler with BLAST as search tool RepeatModeler -engine ncbi -pa 4 -database Pcal_DB >& repeatmodeler_Pcal_assembly.log
Classify files for preperation of the database for RepeatMasker
#classify TE detection RepeatClassifier -engine ncbi -consensi PcalFull_refTEs.fa #classify Hymenoptera repbase RepeatClassifier -engine ncbi -consensi hymenoptera-repbase.fasta
Finally we applied TE-class from online services with default parameter on the repeat library.
Repeat masking with RepeatMasker
RepeatMasker is a program written in perl, which is masking repeats accoding to a repeat library in nucleotide sequences. This programm replaces nucleotides in the sequeces which are included in repetitive sequeces by N's (hard masking) or by lower case characters like a,g,t, and c (soft masking). RepeatMasker uses sequence comparison programs like nhmmer, cross_match, ABBlast/WUBlast, RMBlast and Decypher to identify repetitive sequences in the genome which are also present in the used repeat library (collection of known repeats). For the repeat masking of P.californicus a repeat library was generated by using the repeats from Repbase which are specific for Hymenopterans, the classified repeat families detected by the de-novo approach with RepeatModeler, and the classified transposable elements detected by REPET. In order to increase the completeness of the library and reduce redundancy, we applied TE-class to classify unknown repeats and removed repeats with more then 90 % identity by appling cd-hit.
Run RepatMasker in soft-masking mode on the preperated repeat library ("P.californicus_repeat_library.fa") where the Hymenoptera specific repeats from Repbase, the de-novo detected repeats for P.californicus from RepeatModeler and the detected transposable elements from REPET are included.
#run RepeatMasker nohup RepeatMasker -s -xsmall -a -gff -pa 50 -u -lib P.californicus_repeat_library.fa -dir final_RepeatMasker_out P.cal_polished_new_assembly.fasta >& repeat_annotation.log &
Get final detailed annotation summary.
#get detailed annotation perl buildSummary.pl P.cal_polished_new_assembly.fasta.out > annotation.tbl
Results
Download P.cal repeat library: [P.cal_classified_repeat_library.fa]
Download P.cal RepeatMasker Repeat summary (masked: 20.25 %): [P.cal_RepeatMasker_annotation.tbl]
Download P.cal RepeatMasker detailed Repeat summary: [P.cal_detailed_RepeatMasker_annotation.tbl]
Download P.cal RepeatMasker annotation: [P.cal_RepeatMasker.fasta.gff]
Download P.cal alignment file: [P.cal_RepeatMasker.fasta.align]
Download P.cal RepeatMasker out file: [P.cal_RepeatMasker.fasta.out]
Download P.cal RepeatMasker masked genome assembly fasta file: [P.cal_RepeatMasker.fasta.masked]
References
- György Abrusán, Norbert Grundmann, Luc DeMester, Wojciech Makalowski TEclass—a tool for automated classification of unknown eukaryotic transposable elements , Bioinformatics, Volume 25, Issue 10, 15 May 2009, Pages 1329–1330 https://doi.org/10.1093/bioinformatics/btp084
- Timothée Flutre, Elodie Duprat, Catherine Feuillet, Hadi Quesneville Considering Transposable Element Diversification in De Novo Annotation Approaches, PLOS, Published: January 31, 2011 https://doi.org/10.1371/journal.pone.0016526
- Hadi Quesneville , Casey M Bergman, Olivier Andrieu, Delphine Autard, Danielle Nouaud, Michael Ashburner, Dominique Anxolabehere Combined Evidence Annotation of Transposable Elements in Genome Sequences , PLOS, Published: July 29, 2005 https://doi.org/10.1371/journal.pcbi.0010022