Institute of Bioinformatics Münster
Assembly Summary
Summary of assemblies
Description
This page was created to store the assembly data that was used for further analysis. There are two tables describing different types of sequence assemblies (genome assemblies and transcript assemblies). The Overview section describes the transcriptome construction.Genome Data
| Species | Download | Assembly level | Sequencing method | Material | Assembly Tool | Number of sequences | Nucleotides in sequences [nt] | Shortest sequence [nt] | Longest sequence [nt] | Median sequence length | Mean sequence length | N50 [nt] | NG50 [nt] | N90 [nt] | Percentage of N's | Total number of N's | GC-content | Sequence size distribution graph | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pogonomyrmex californicus | [Supernova 2.1 assembly] [Download fasta index] |
Scaffold | 10X Genomics | nuclear DNA | Supernova 2.1 | 6,793 | 241,082,378 | 460 | 1,993,378 | 5,353 | 35,489 | 208,871 | 1,993,378 | 16,229 | 1.15 % | 3,132,150 | 36.3 % |
[View part] |
- |
| Pogonomyrmex californicus | [Supernova 2.0 assembly] [Download fasta index] |
Scaffold | 10X Genomics | nuclear DNA | Supernova 2.0 | 12,983 | 314,017,594 | 395 | 8,422,444 | 4,713 | 24,187 | 1,497,864 | 8,422,444 | 9,161 | 3.07 % | 9,650,154 | 36.6 % |
[View global] [View detailed] |
- |
| Pogonomyrmex barbatus | - | Scaffold | 454 XLR titanium platform at SeqWright | nuclear DNA | CABOG (v. 5.3) | 4,645 | 235,645,958 | 297 | 3,918,663 | 1,353 | 50,731 | 819,605 | 3,918,663 | 117,988 | 6.6 % | 15,575,506 | 34 % |
[View global] [View detailed] |
RefSeq assembly accession: GCF_000188075.1 NCBI ID: 243358 |
| Camponotus floridanus | - | Scaffold | PacBio | nuclear DNA | canu (v. 1.3) | 10,791 | 284,009,182 | 6,425 | 10,163,455 | 106,195 | 432,281 | 1,585,631 | 10,163,455 | 211,219 | 2.87 % | 8,162,210 | 26 % |
[View global] [View detailed] |
RefSeq assembly accession: GCF_003227725.1 NCBI ID: 1752781 |
| Solenopsis invicta (red fire ant) | - | Scaffold | 454 Illumina | nuclear DNA | SOAPdenovo (v. 1.04) Newbler (v. 2.3) | 69,511 | 396,024,718 | 200 | 6,355,204 | 721 | 5,967 | 558,018 | 6,355,204 | 12,516 | 0.13 % | 538,573 | 25 % |
[View global] [View detailed] |
RefSeq assembly accession: GCF_000188075.1 NCBI ID: 244018 sex: male |
| Solenopsis invicta | - | Chromosome | 454 Illumina | mitochondrial DNA | SOAPdenovo (v. 1.04) Newbler (v. 2.3) | 1 | 15,549 | - | - | - | - | - | - | - | 0 % | 0 | 22.83 % | - | RefSeq assembly accession: NC_014672.1 |
| Apis mellifera (honey bee) | - | Chromosome | Sanger SOLiD 454 | nuclear DNA | Atlas assembly system (v. before 2011) | 5,321 | 250,287,000 | 540 | 29,893,408 | 1,351 | 47,037 | 13,219,345 | 29,893,408 | 147,519 | 0.11 % | 276,584 | 21.67 % |
[View global] [View detailed] |
RefSeq assembly accession: GCF_000002195.4 NCBI ID: 254398 |
Transcriptome Data
| Species | Download | Assembly level | Sequencing method | Source | Assembly Tool | Number of sequences | nucleotides in sequences [nt] | shortest sequence [nt] | longest sequence [nt] | Median sequence length | Mean sequence length | N50 [nt] | NG50 [nt] | N90 [nt] | percentage of N's | total number of N's | GC-content | Sequences size distribution graph | Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pogonomyrmex californicus | [Downloaded from here] | transcript contigs | Illumina Hiseq 2000 [Done by M. Helmkampf et al. (2016)] |
RNA from Head | Trinity (v. 1) [Done by M. Helmkampf et al. (2016)] |
7,890 | 31,829,855 | 237 | 27,920 | 3,434 | 4,034 | 5,055 | 27,920 | 2,269 | 0 % | 0 | 38.2 % |
[View detailed] [View global] [View filtered global] [View filtered detailed] |
This assembly was published by M. Helmkampf et al. (2016), trimmed with Trim_galore v. 0.3.1 and filtered by FPKM of 0.14. |
| Pogonomyrmex californicus |
[Download fasta] [Download gff] |
transcript contigs | Illumina Hiseq 2000 [Done by M. Helmkampf et al. (2016)] |
RNA from Head | Trinity (v. 2) | 160,741 | 137,321,817 | 251 | 19,115 | 547 | 854 | 1,219 | 19,115 | 360 | 0 % | 0 | 37.7 % |
[View detailed] [View global] [View filtered global] [View filtered detailed] |
Used published RNA raw reads. Trimmed with Trim_galore v. 0.5.0. |
| Pogonomyrmex californicus |
[Download fasta] [Download gtf] |
transcript contigs | Illumina Hiseq 2000 [Done by M. Helmkampf et al. (2016)] |
RNA from Head | Tuxedo Suite (v. 1) | 34,953 | 71,805,488 | 79 | 26,134 | 1,490 | 2,054 | 2,760 | 26,134 | 1,005 | 0.004 % | 2,772 | 37 % |
[View global] [View detailed] |
Used published RNA raw reads.Trimmed with Trim_galore v. 0.5.0: |
| Pogonomyrmex californicus |
[Download fasta] [Download gtf] |
transcript contigs | Illumina Hiseq 2000 [Done by M. Helmkampf et al. (2016)] |
RNA from Head | Tuxedo Suite (v. 2) | 51441 | 107,364,332 | 200 | 30,234 | 1,513 | 2,087 | 3,163 | 30,234 | 1,033 | 0.002 % | 2,741 | 37.64 % |
[View global] [View detailed] |
Used published RNA raw reads.Trimming with Trim_galore v. 0.5.0: |
| Pogonomyrmex californicus |
[Download fasta] [Download gtf] |
transcript contigs | MinION | RNA | - | 10,700 | 12,715,775 | 200 | 31,833 | 991 | 1,188 | 1,450 | 31,833 | 648 | 0 % | 0 | 35.49 % |
[View global] |
Stringtie (v. 2.0) |
| Pogonomyrmex californicus |
[Download fasta] [Download gtf] |
merged transcript assembly | Illumina and MinION | RNA | Stringtie (v. 2.0), Tuxedo Suite (v. 1 and v. 2) and Trinity (v. 1 and v. 2) |
31,515 | 52,754,802 | 200 | 33,094 | 1,171 | 1,673 | 2,502 | 33,094 | 812 | 0.03 % | 18,348 | 37.4 % |
[View global] [View detailed] |
Merged final transcript assembly (see Overview) |
Discussion
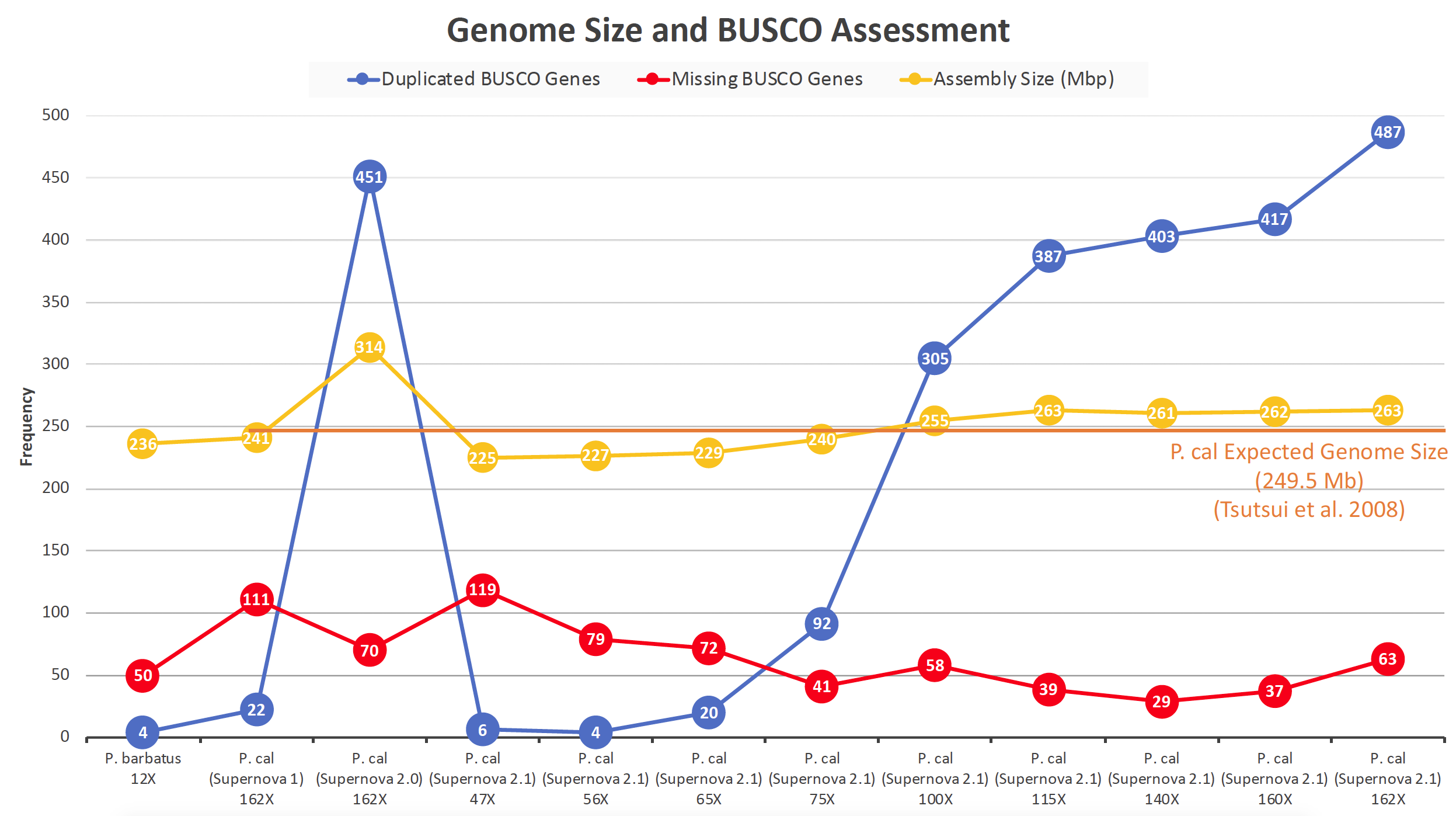
- Figure 1: Evaluation of the quality and completeness of genome assemblies from different species and assemblers. The P. californicus assemblies have been assembled with Supernova v. 1, 2.0, and 2.1. The assembler Supernova (v. 2.1) includes the option of choosing a coverage. Assemblies with different genome coverage have been generated and evaluated.
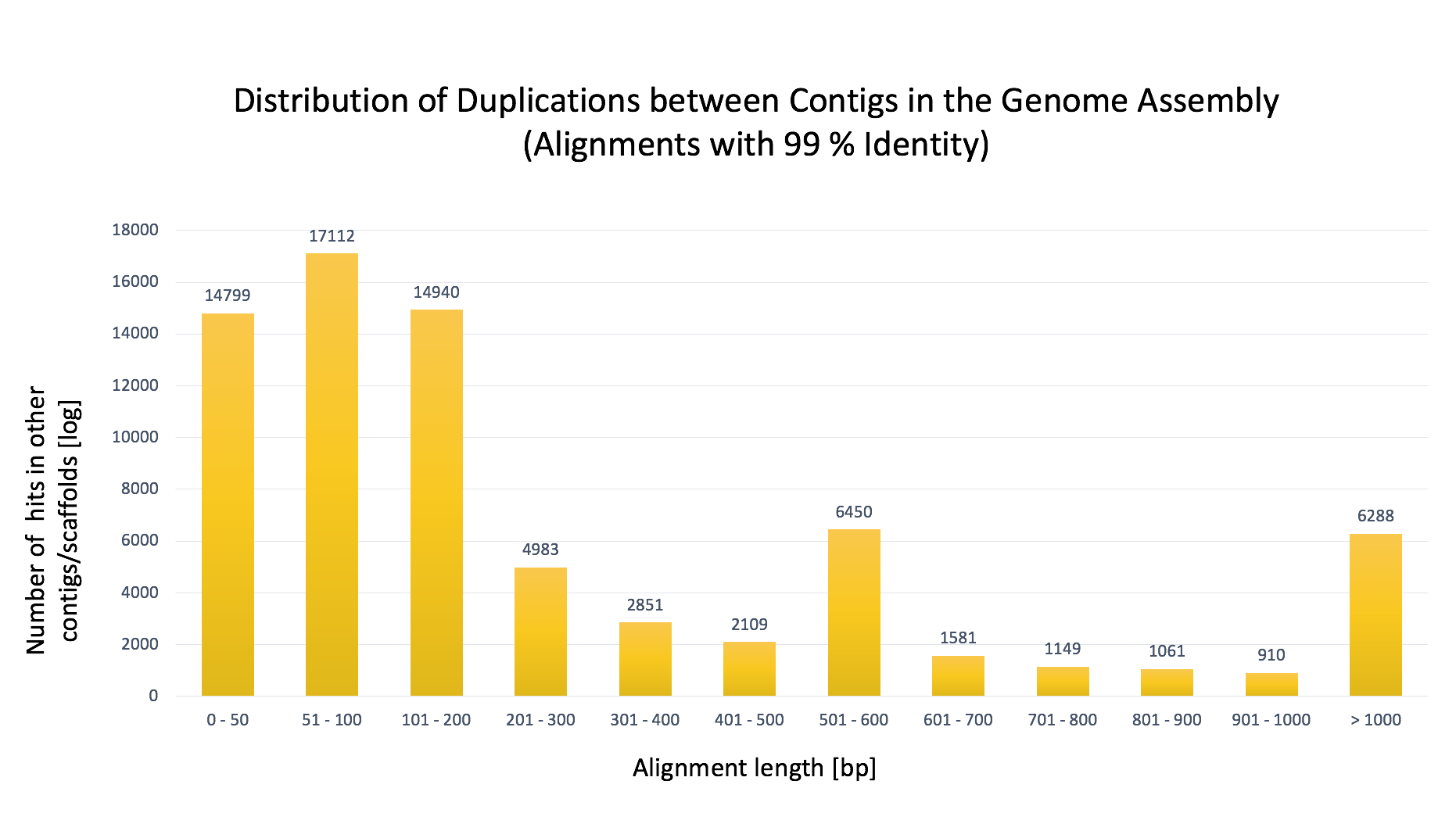
- Figure 2: Distribution of inter contig duplications within the Supernova 2.1 (75x) genome assembly.
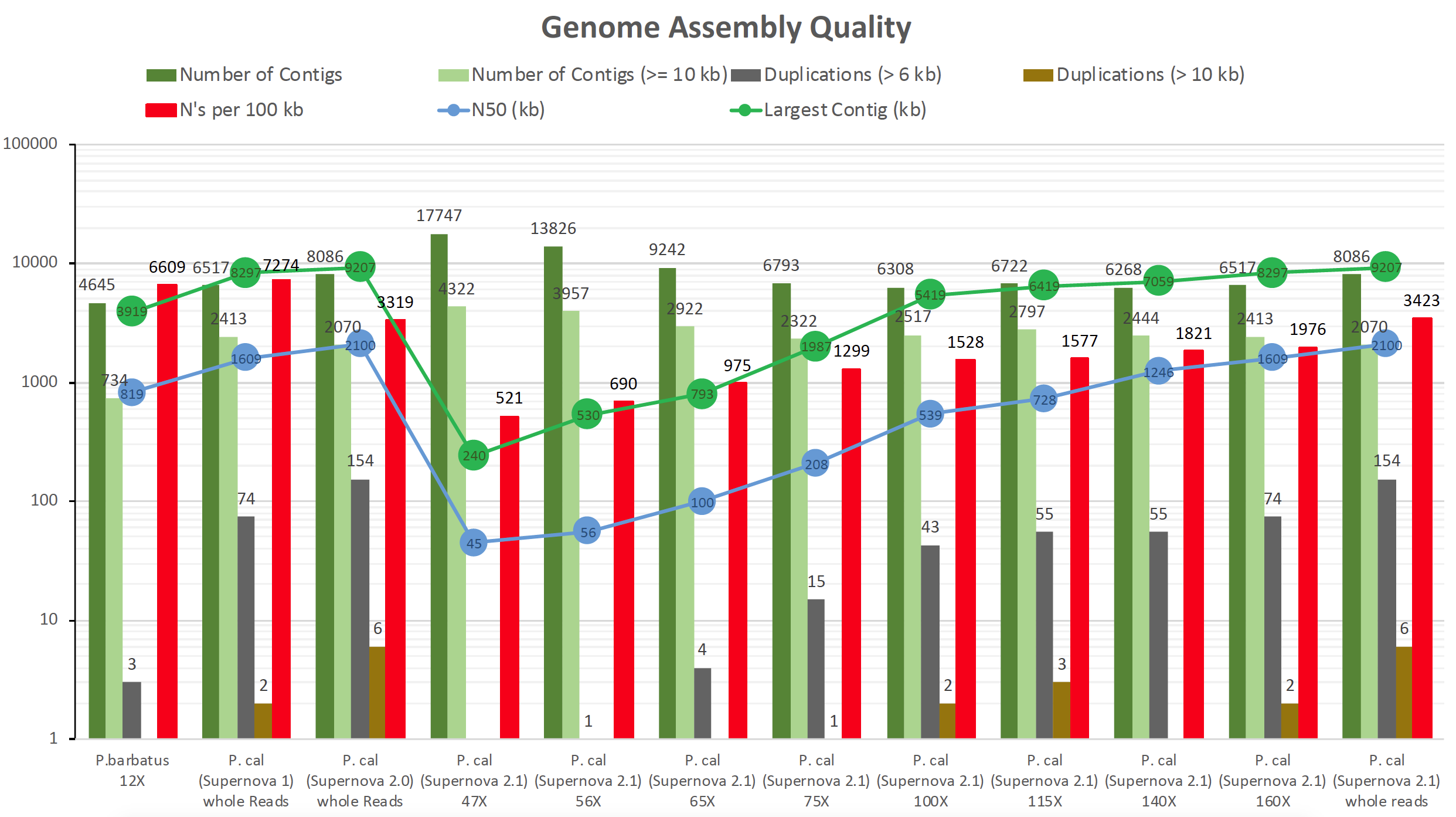
- Figure 3: Detailed Plot for displaying a higher resolution of duplicated sequences within the genome assemblies from Figure 1.
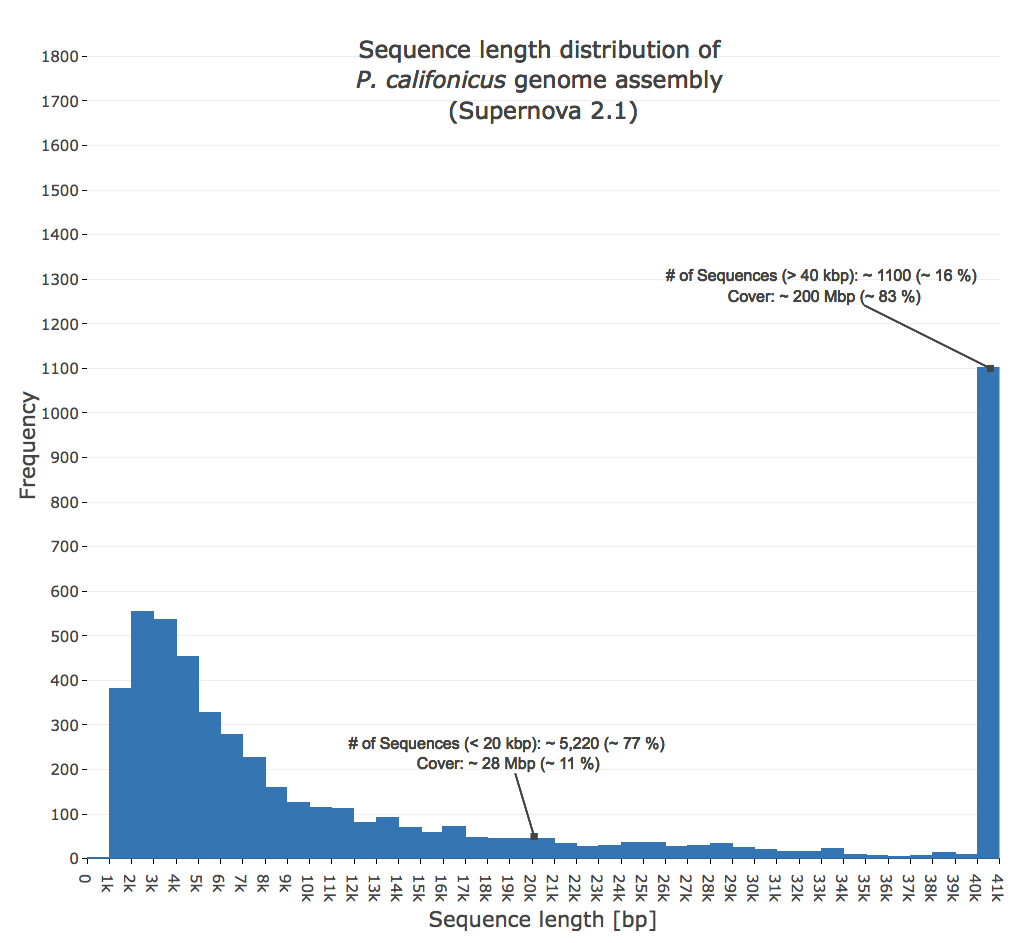
- Figure 4: Genome assembly sequence length distribution. Based on the genome assembly which was generated by Supernova 2.1 with the sequencing depth of 75x.The last bar in this graph displays all sequences which are longer than 40 kbp.
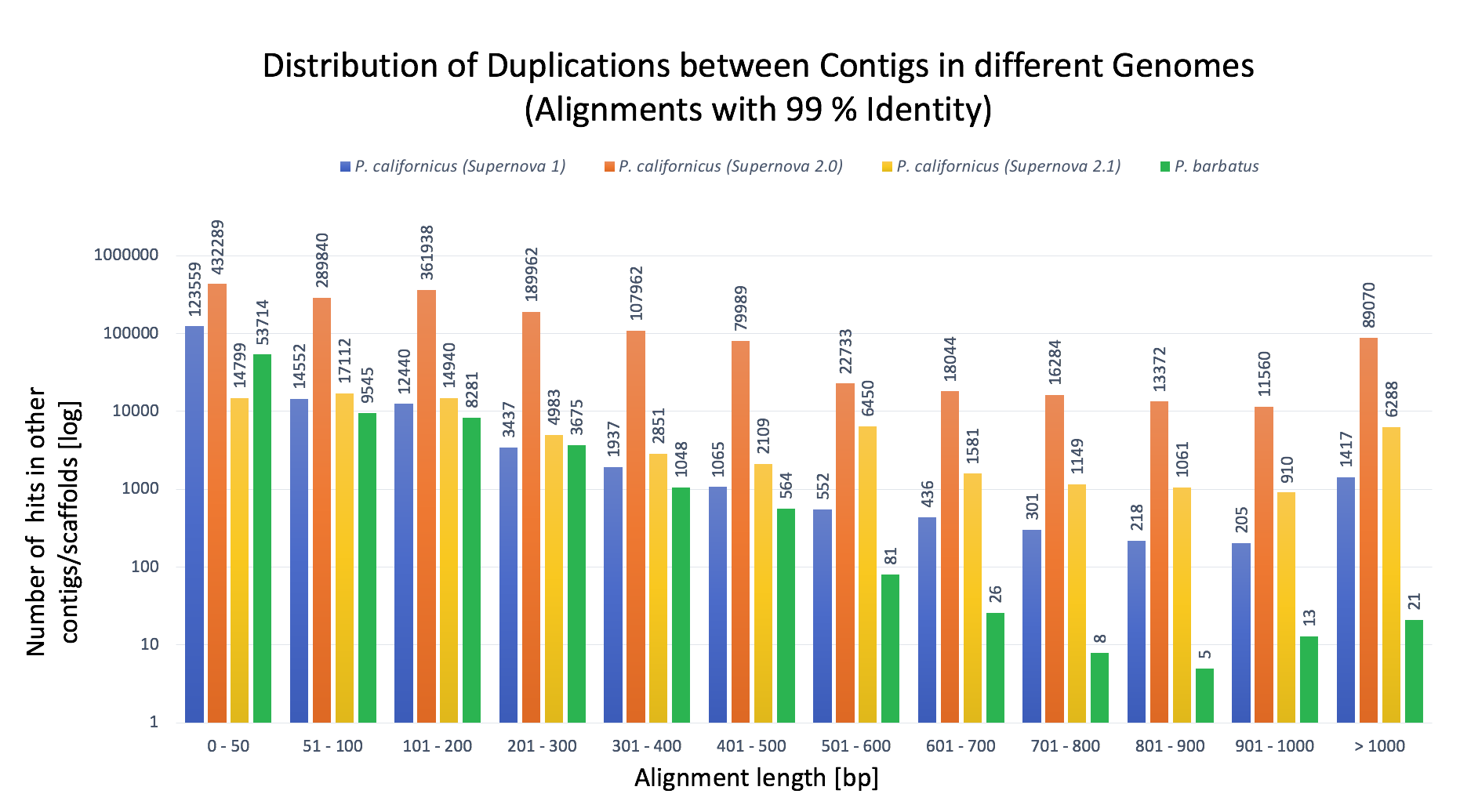
- Figure 5: Detailed Plot for displaying duplicated sequences within the genome assemblies between contigs.
2020-11-18 19:53